Desirazu N. Rao

Desirazu N. Rao
Honorary Professor
Ph.D.: Indian Institute of Science, Bangalore, India
Post-doctoral research:
National Institutes of Health, USA, Biozentrum, Switzerland, University of Cambridge, U.K.
Year of Joining: 1989
Email:dnrao@iisc.ac.in
We study DNA-Protein interactions using Helicobacter pylori Restriction-Modification enzymes and Neisseria gonorrhoea DNA Mismatch Repair proteins as model systems.
Highlights
- Both C5 and N4- methyl cytosine signals act as global epigenetic regulators in H. pylori
- Phase variable property of DNA methylases contribute to variability
- H pylori Topoisomerase I has distinct properties, in that the C-terminal domain alone comprising zinc finger motifs show DNA relaxation activity
Helicobacter pylori
Helicobacter pylori strains are rich in Restriction- Modification (R-M) systems and several genes encoding these R-M systems are strain specific. These systems are vital in the balance between genetic diversity and genomic integrity of this pathogen. DNA methyltransferases epigenetically regulate the expression of many genes through the precise epigenetic marking of certain cytosines or adenines with a methyl group.
Research from our laboratory has shown that deletion of an orphan cytosine C5 DNA methylase in H. pylori strains alters the expression of genes involved in motility, adhesion and virulence. A few H. pylori strains harbor a single N4 cytosine methyltransferase which generates an m4C signal on DNA. Recent work from our laboratory revealed the role of m4C modification by deleting the gene in H. pylori 26695. Upon the loss of m4C modification, a total of 102 genes belonging to virulence, ribosome assembly and cellular components were differentially expressed. Our study provides the first evidence that m4C signal acts as a global epigenetic regulator in H.pylori.
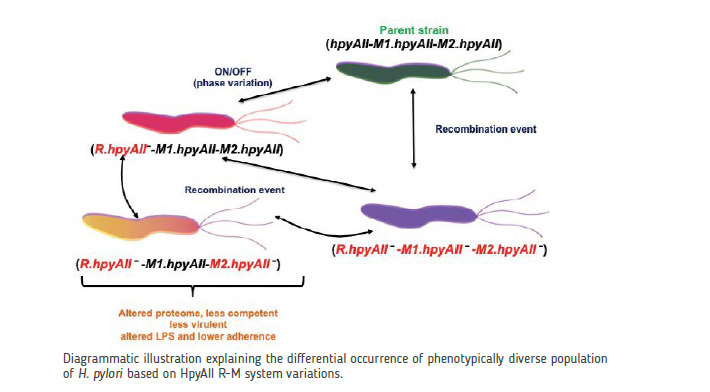
N6-adenine methylases modify the adenine to generate an m6A epigenetic signal. We have shown that a phase-variable N6 adenine DNA methylase in H. pylori is highly conserved in all known strains. The expression of the active protein in a strain is dependent on the phase variation status at a homopolymeric repeat locus within the gene. Preliminary results suggest that the methylation status dictated by this protein in H. pylori 26695 is important for outer membrane function and stress-response.
Topological homeostasis is maintained in the cells by incorporation or removal of supercoils to alleviate the strain on the DNA. Topoisomerases are ubiquitous molecular scissors whose characteristic property is to preserve the DNA topology during various cellular processes. A regulatory role of H. pylori topoisomerases in altering DNA supercoiling and thus controlling gene expression has not been studied. The essentiality of the two topoisomerases in H. pylori makes them druggable targets to circumvent infection. Work from our lab explores Topoisomerase I function via extensive biochemical characterization. We have revealed that topoisomerase I has features that is different from homologues in other bacteria, pathogenic or otherwise. We have elucidated the roles of the amino terminal Toprim domain and carboxyl terminal zinc finger motifs in DNA binding and enzyme catalysis. We believe the compact genome of H. pylori may have led to a directed evolution pathway by which a smaller topoI enzyme, the C-terminal domain alone, is capable of catalysis.
Although recent evidence suggests that acetylated proteins broadly impact cellular functions in prokaryotes, the substrates and localization of this modification in H. pylori or N. gonorrhoeae remain widely unknown. We compared the acetylome profile of various H. pylori strains using anti-acetylated lysine antibody. There was a prominent acetylation in each strain as well as significance difference between their acetylation patterns. Sequence analysis of known protein acetyltransferases from different bacteria against H. pylori 26695 genome identified Hp0935 as a putative protein N-acetyltransferase. Crystal structure of Hp0935 shows that it has GNAT fold. HpDprA (DNA processing protein A) and M1.HpyAVI were found to be acetylated by Hp0935 (enzymatically) as well as non-enzymatically in-vitro. These findings suggest a potential role of acetylation in H. pylori competency.
Neisseria gonorrhoeae
The mismatch repair (MMR) system maintains the integrity of the genome primarily by removing any non-Watson Crick base pairs that arise due to mis-incorporation during replication. N. gonorrhoeae does not possess any homologs of MutH and therefore the strategy utilized for strand discrimination and creation of a nick in the daughter strand to initiate MMR will be different in this organism. The structure of the C-terminal domain of N. gonorrhoeae MutL solved (in collaboration with Deepak Nair’s lab at RCB, Faridabad) showed that the protein exists in the form of an elongated inverted homodimer that presents each active site on the lateral faces of the molecule. UvrD helicase is another integral component of the MMR pathway. However, we have recently demonstrated that N. gonorrhoeae UvrD can unwind G-quadraplex structures and may have an important role in phase variation of Type IV pilus system in this organism which contains a G-rich region that can form G-quadraplex structures.
- Probing of interactions between proteins from bacterium Neisseria gonorrhoeae mismatch repair system by mass spectrometric analysis of crosslinked biomolecules; Indo-Russian Joint Project; 2017-2019;
- Epigenetic regulation by phase-variable Type IIs R-M system of Helicobacter pylori; Department of Biotechnology; 2017-2020;
- Molecular interactions critical for DNA mismatch repair; Department of Science & Technology; 2017-2020;
- Identification of virulence-associated genes of H. pylori from understudied South Indian population of gastric cancer and gastritis; RGUHS, Bangalore; 2018-2020;
- Narayanan, N., Banerjee, A., Jain, D., Kulkarni, D.S., Sharma, R., Nirwal, S., Rao, D.N., and Nair, D.T. (2019). Tetramerization at low pH licenses DNA methylation activity of M.Hpy A XI in the presence of acid stress. J. Mol. Biol. (in press)
- Damke, P.P., Di Guilmi, A.M., Charbonnier, JB., Marsin, S., Veaute, X., Gunjal, G.V., Rao, D.N. and Radicella, J.P. (2019). Identification of the periplasmic DNA receptor for natural transformation of Helicobacter pylori. Nat. Commun. (in press)
- Prasad, Y., Kumar, R., Chaudhary, A. K., Dhanaraju, R., Majumdar, S., and Rao, D. N. (2019) Kinetic and catalytic properties of M.Hpy A XVII, a phase-variable DNA methyltransferase from Helicobacter pylori. J. Biol. Chem. 294, 1019–1034
- Nirwal, S., Kulkarni, D. S., Sharma, A., Rao, D. N. and Nair, D. T. (2018) Mechanism of formation of atoroid around DNA by the mismatch sensor protein. Nucleic Acids Res 46,256-266
- Kumar, S., Karmakar, B. C., Nagarajan, D., Mukhopadhyay, A. K., Morgan, R. D. and Rao, D. N. (2018) N4-cytosine DNA methylation regulates transcription and pathogenesis in Helicobacter pylori. Nucleic Acids Res. 46, 3429–3445
Ph.D. Students 3
Undergrads 0
Postdocs 1
Trainees 2