Ganesh Nagaraju

Ganesh Nagaraju
Professor
Ph.D.: Indian Institute of Science, Bangalore, India
Post-doctoral research:
Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, USA
Year of Joining: 2007
Email: nganesh@iisc.ac.in
Our lab is broadly interested in understanding how various genes regulate DNA damage responses, maintain genome integrity and supress tumorigenesis.
Highlights
- ATR signaling distinctly activate XRCC2 and XRCC3 for genome maintenance and cell survival
- RAD51 paralogs protect and restart stalled replication forks
- RAD51 paralogs facilitate mitochondrial DNA replication and maintain integrity of mitochondrial genome
Mammalian genome encodes five RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2 and XRCC3). We have established that RAD51C is a novel gene in the Fanconi anemia (FA) pathway of DNA interstrand cross-link (ICL) repair and demonstrated that RAD51C distinctly regulates intra-S-phase checkpoint and DNA repair. This finding has implications for FA and breast and ovarian cancer susceptibility. We have demonstrated that XRCC3 is a novel phosphorylation target of ATM and ATR kinases which is crucial for execution of DNA repair and intra-S-phase checkpoint. We showed new roles of RAD51 paralogs in the protection and restart of stalled forks. Our study with pathological mutants of RAD51C provided insights into tumor suppressor and essential functions of RAD51 paralogs in genome maintenance. In collaboration, we have developed a novel photo-inducible DNA ICL molecule for cancer therapy. Our work showed roles of RAD51 paralogs beyond its nuclear functions in facilitating mitochondrial DNA replication and in maintaining mitochondrial genome stability. We find that XRCC2 restrains pathological fork progression during dNTP alterations (Fig. 1). XRCC2 and XRCC3 are differentially activated by ATR kinase to protect the genome from DNA lesions and cell death. We also identified novel function of FANC-Jheli case in suppressing gene amplification.
Mycobacterium tuberculosis genome is GC rich (65%) and there are >10,000 G-rich motifs that have the potential to form G4 structures. We identified DinG as a G4 DNA resolving helicase in mycobacteria. Our work also demonstrated that M. tuberculosis RecGheli case drives efficient reversal of stalled for ks which has implications for replication restart mechanisms in M. tuberculosis.
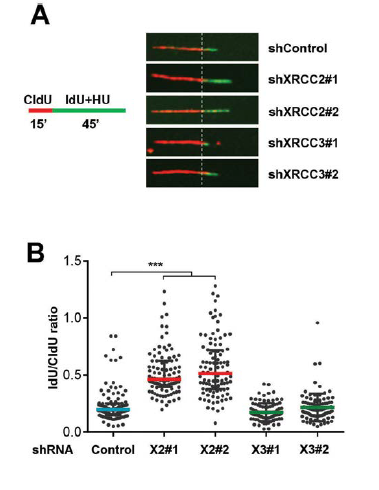
FIG 1 : XRCC2 is required for efficient fork slowdown during nucleotide pool perturbations. (A) Representative set of DNA fibers to display fork slowing induced by HU (500 μM) in U2OS cells treated with indicated shRNAs #1 and #2 indicate two independent shRANs. (B) idU to CldU ratios of fibers from cells as shown in (A).
Future Plans
Homologous recombination (HR) and translesion synthesis (TLS) contribute to DNA damage tolerance in mammalian cells. However, it is unclear how the seprocesses are regulated. Currently, we are investigating whether RAD51 paralogs fine tune HR and TLS pathways to maintain integrity of the mammalian genome and supress tumorigenesis. Our preliminary studies demonstrate that FANCJ helicase controls resection of DSB ends. We wish to study the molecular mechanism by which FANCJ heli case participates in DSB repair. We also wish to understand the role of BLM and RTEL helicases in recombinational repair of DSBs and their pro- and anti-recombination functions in genome maintenance. DNA damage induced spontaneously or by exogenous our ces hampers DNA replication and, replication associated DNA lesions are the major source of genome instability that cause tumor igenesis. Cells have evolved with different pathways to deal with replication problems and there by maintain integrity of the genome. Our long term goal is to study these pathways and mechanisms in mammalian cells.In addition, studies are under progress to uncover new gene sand their mechanisms that contribute to the integrity of mammalian genome and suppress tumor igenesis.Results obtained from these studies would be used to translate this knowledge to develop novel therapeutic strategies for the treatment of genetic diseases and cancer.
- Understanding the DNA damage tolerance pathways and mechanisms in mammalian cells; Department of Biotechnology; 2018-2021;
- Role of RAD51C and XRCC3 mediated chromatin remodeling in the regulation of DNA repair and DNA damage signaling, Department of Biotechnology-National Bioscience Award; 2017- 2020;
- Understanding the role of RTEL1 helicase at the sites of damaged replication forks; Department of Science and Technology; 2016-2019;
- Regulation of gene conversion and crossover control by Fanconi anemia pathway FANCJ helicase; Department of Biotechnology; 2014- 2017;
- Nath, S. Nagaraju, G. (2020). FANCJ helicase promotes DNA end resection by facilitating CtIP recruitment to DNA double-strand breaks. PLoS Genetics. 16(4): e1008701.
- Saxena, S., Dixit, S., Somyajit. K, and Nagaraju, G. (2019). ATR signaling uncouples the role of RAD51 paralogs in homologous recombination and replication stress response. Cell Rep. 29: 551-559
- Saxena, S., Somyajit, K. and Nagaraju, G. (2018). XRCC2 regulates replication fork progression during dNTP alterations. Cell Rep. 25: 3273-3282
- Mishra, A., Saxena, S., Kaushal, A. and Nagaraju, G. (2018) RAD51C/XRCC3 facilitates mitochondrial DNA replication and maintains integrity of the mitochondrial genome. Mol. Cell. Biol. 38,1-18
- Nath, S., Somyajit, K., Mishra, A., Scully, R. and Nagaraju, G. (2017) FANCJ helicase controls the balance between short- and long-tract gene conversions between sister chromatids. Nucleic Acids Res. 45,8886-8900
- Somyajit, K., Saxena, S., Babu, S., Mishra, A. and Nagaraju, G. (2015) Mammalian RAD51 paralogsprotect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res. 43,9835-9855
Ph.D. Students 7
Postdocs 2
Undergrads 0
Trainees 1