Sandeep M. Eswarappa

K. Muniyappa
CSIR Bhatnagar Fellow & Honorary Professor
Ph.D.: Indian Institute of Science, Bangalore, India.
Post-doctoral research:
Yale University School of Medicine, New Haven, USA University of Georgia, Athens, USA
Year of Joining: December 23, 1987
Email:kmbc@iisc.ac.in
Research in my laboratory is focused on understanding the mechanisms of chromosome synapsis, homologous recombination, DNA damage recognition and repair. A multi-disciplinary approach is employed using molecular cell biological to structural biology to single molecule imaging of protein-DNA interactions of biochemical reactions.
Highlights
- Discovered a negative regulatory mechanism of RecA promoted homologous recombination by RecX, second messenger cyclic-di AMP and anionic phospholipids.
- Provided the first direct evidence that DNA damage response proteins are essential for telomere length homeostasis, cell senescence and cell cycle checkpoint control.
- Has made pioneering contributions in bringing higher complexity to the biochemical issues of cellular recombination and resolution of the Holliday junction intermediates.
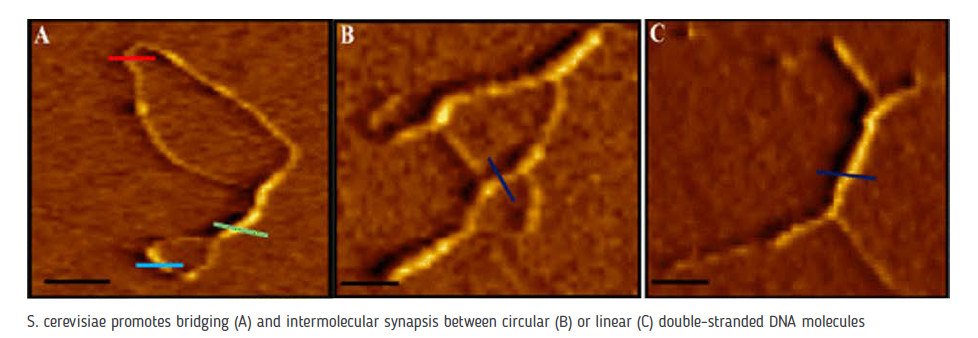
Biochemical and structural analysis of Saccharomyces cerevisiae synaptonemal complex (SC) proteins.Meiosis is a specialized form of cell division during which the diploid cells undergo a single round of DNA replication followed by two rounds of chromosome segregation to produce haploid gametes. At the center of this process is the SC, a dense, evolutionarily conserved, proteinaceous superstructure that functions as a “molecular zipper” to juxtapose homologous chromosomes, one from each parent, to pair up and exchange genetic material by crossing over. A growing body of evidence suggests that defects in the SC leads to infertility, recurrent miscarriages and chromosome aneuploids such as the Down’s syndrome, in addition to germline cancers. How the components of SC exert their function has been hotly debated from multiple perspectives. Our research has focused on delineating the structure-function relationships of S. cerevisiae SC proteins, the functional relationships among them, and the underlying molecular mechanism(s) by which these proteins may orchestrate meiotic chromosome synapsis and recombination. We have employed an ensemble of molecular biological and single molecular approaches to characterize the activities of Hop1 and Red1, the two meiosis-specific proteins of the SC axial element of S. cerevisiae.
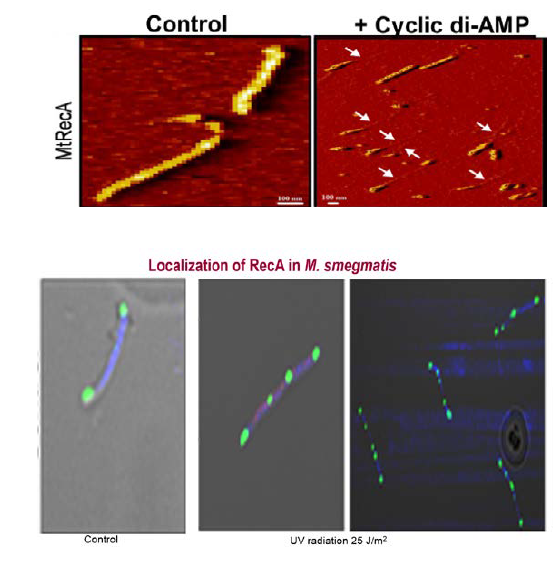
Physiologically important small molecules as regulators of RecA function: The bacterial RecA protein plays a pivotal role in the repair of stalled replication forks, double-strand break repair, homologous recombination, and the SOS response. RecA is regulated at many levels. The biological activities of RecA protein is regulated by the action of a number of accessory proteins, including RecF, SSB, RecO, RecR, DinI, RecX, RdgC, PsiB, and UvrD. All of these proteins typically exert their actions on the assembly and/or activity of RecA nucleoprotein filaments through positive or negative regulatory mechanisms. To further inform our understanding of the regulation of RecA function by endogenous small molecules, the potential roles of the nucleotide second messengers and plasma membrane components were examined. Our work support the notion that cyclic di-AMP, but not cyclic di-GMP, and anionic phospholipids of the bacterial plasma membrane regulate recombination-based DNA repair and SOS response.
- Center of Excellence on genetic recombination in mycobacteria.
- CSIR Bhatanagar Fellowship
- 1. Kshirsagar, R., Ghodke, I., and Muniyappa, K. (2017) Saccharomyces cerevisiae Red1 protein exhibits nonhomologous DNA end-joining activity and potentiates Hop1-promoted pairing of double-stranded DNA. The Journal of Biological Chemistry 292, 13853-13866
- Kshirsagar, R., Khan, K., Joshi, M. V., Hosur, R. V., and Muniyappa, K. (2017) Probing the Potential Role of Non-B DNA Structures at Yeast Meiosis-Specific DNA Double-Strand Breaks. Biophysical Journal 112, 2056-2074
- Thakur, M., Kumar, M. B., and Muniyappa, K. (2016) Mycobacterium tuberculosis UvrB Is a Robust DNA-Stimulated ATPase That Also Possesses Structure-Specific ATP-Dependent DNA Helicase Activity. Biochemistry 55, 5865-5883
- Kaulage, M., Maji, B., Bhat, J., Iwasaki, Y., Chatterjee, S., Bhattacharya, S., and Muniyappa, K. (2016) Discovery and Structural Characterization of G-quadruplex DNA in Human Acetyl-CoA Carboxylase Gene Promoters: Its Role in Transcriptional Regulation and as a Therapeutic Target for Human Disease. Journal of Medicinal Chemistry 59, 5035-5050
- Nautiyal, A., Rani, P. S., Sharples, G. J., and Muniyappa, K. (2016) Mycobacterium tuberculosis RuvX is a Holliday junction resolvase formed by dimerisation of the monomeric YqgF nuclease domain. Molecular Microbiology 100, 656-674
- Manikandan, K.,® Prasad, D.,® Srivastava, A., Singh, N., Dabeer, S., Krishnan, A., Muniyappa, K.© and Sinha, K. M. © (2018) The second messenger cyclic di-AMP negatively regulates the expression of Mycobacterium smegmatis recA and attenuates DNA strand exchange through binding to the C-terminal motif of mycobacterial RecA proteins. Molecular Microbiology 109(5):600-614. ® Equal first authors; ©Co-corresponding authors.
List of Ph.D students
- Somashekhar Gowda
- Supreet Bhattacharya
- Ankit Agarwal
- Kshitiza Mohan Dhyani
List of Post Docs
- Sugith Badugu
- Akhilesh Kushawaha
- P. Sanjeev