Mahipal Ganji

Mahipal Ganji
Assistant Professor
Ph.D: TU Delft, The Netherlands
Post-doctoral research:
TU Delft, The Netherlands and Max-Planck Institute of Biochemistry Martinsried, Germany
Year of Joining: 2020
Email: ganji@iisc.ac.in
We are a multidisciplinary research group interested in understandingthe macromolecular interactionsthat help in 3D genome organization. In this regard, we develop and employ single-molecule and super-resolution imaging technologies.
Highlights
- Non-covalently bound macromolecular complexes undergo rapid re-bindings.
- First proof for DNA loop-extrusion by SMC complexes.
- Super-resolution imaging with transient interactions of short peptides.
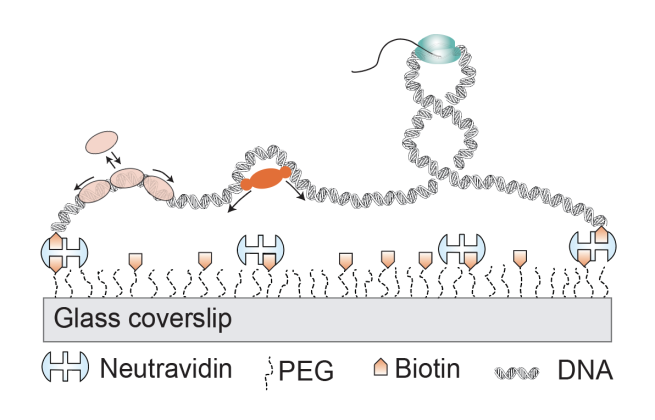
Single-molecule assay to visualize the structure of DNA in response to DNA binding proteins
Single-molecule visualization of DNA-protein interactions: In the past, we developed single-molecule assay to visualize the dynamics of supercoiled DNA based on Total Internal Reflection Fluorescence microscopy. We made use of intercalating dye to introduce supercoiling on a surface bound DNA molecule.Using this assay, we figured out that intrinsically bent DNA – due to local sequence – facilitates for positioning the intertwined DNA structure called plectoneme (Ganji et al, Nano Letters, 2016, Kim et al, Elife, 2018). Furthermore, using this assay we also studied how SMC (Structural maintenance of chromosomes) proteins introduce loops into DNA. We recorded real-time videos to visualize the action of SMC proteins on surface tethered DNA molecules to show that SMC proteins use active loop-extrusion mechanism (Ganji et al, Science, 2018).
In our future experiments, we focus to understand the mechanism of DNA compaction and protection by nucleoid associated proteins (NAPs) in bacteria.We will probe the effect ofDNA supercoiling on transcription using the supercoiled DNA assay (Ganji et al, Nano Letters, 2016). Furthermore, we will study how transcription proceeds in the presence of DNA compacting proteins. In addition, we will develop tools to visualize DNA processingenzymesfor extended periods while they are performing their function. This will allow us to answer several questions pertaining to target search mechanism, interactions of multitude of DNA binding proteins at single-molecule level.
Genome organization: Human genome is compacted by five orders of magnitude to fit a 2-meter long genome in micron sized compartment, yet the active and inactive chromatin domains tend to spatially segregate. This compartmentalizationhappens mainly due to the posttranslational modifications on the histone tails, also known as epigenetic marks. We are interested in understanding the interplay between different epigenetic marks in genome compaction and spatial distribution of individual genes. Our aim is to map the 3D organization of active genes and their transcriptional activity at single-cell level. For this, we will develop multiplexed quantitative imaging approaches using DNA-PAINT super-resolution microscopy.
These multiplexed super-resolution assays will also be applied to image the higher order chromatin organization based on the epigenetic marks in different cell types and disease variants.
Will be updated shortly
- Eklund A. et al Peptide-PAINT Super-resolution imaging using transient coiled-coil interactions, Nano Letters, Aug 2020
- Ganji M. et al Real-time imaging of DNA loop extrusion by condensin, Science, 2018
- Kim S. H. et al DNA sequence encodes the position of DNA supercoils, elife, 2018
- Ganji M. et al DNA binding proteins explore multiple local configurations during docking via rapid rebinding, Nucleic Acids Research, 2016
- Ganji M. et al Intercalation-based single-molecule fluorescence assay to study DNA supercoil dynamics, Nano Letters, 2016
Openings