Nagasuma Chandra

Nagasuma Chandra
Professor
Ph.D.: University of Bristol, UK
Post-doctoral research:
Indian Institute of Science, Bangalore
Year of Joining: 2011
Email:nchandra@iisc.ac.in
A major interest in the laboratory is to understand genome-wide perturbations in several human diseases using multi-level modelling of biological systems and translate such insights into biomarker and drug discovery. A specific focus is to understand how anti-microbial resistance emerges and identify new strategies and drug-combinations for tackling resistance.
Highlights
- MDR and XDR-TB: Identification of the Achilles’ heel and a new drug-combination
- TB diagnosis: Discovery of a new host-gene biomarker signature from patient blood samples
In the last few years, my laboratory has been working on obtaining a molecular systems view of Mycobacterium tuberculosis (Mtb) with an aim of (a) understanding mechanisms of drug resistance in the pathogen and discovering strategies for tackling drug resistant varieties, (b) probing genomescale metabolic networks in the pathogen under different stress conditions and identifying the set of essential genes, (c) identifying molecular networks perturbed in macrophages and whole blood samples from tuberculosis patients to identify maximally responding modules and host biomarkers based on them, (d) understanding iron homeostasis in infection and (e) obtaining a structural view of the mycobacterial proteome, characterize its pocketome and identifying druggable targets as well as repurposable drug candidates.
Systems biology of drug resistant mycobacteria: Drug resistance poses a major threat to the successful treatment of tuberculosis. Bacteria are known to have multiple ways of overcoming the drug stress and the complex interactions among the many molecular players warrant a systems view. The specific questions, we have addressed are (i) What are the molecular determinants of isoniazid-resistance in mycobacteria, (ii) Does the resistant strain exhibit differences in its growth fitness, and (iii) Does the resistant strain have an Achilles’ heel that can be identified and used for selecting co-targets that can be inhibited as a new strategy for tackling resistance. These were addressed by first generating genomics data, integrating them onto genome-scale molecular networks, and then applying sensitive network mining approaches. The hypotheses obtained from these were tested by a collaborator Dr. Amit Singh, IISc, by in-vitro and infection experiments. We find multiple resistance mechanisms are operative simultaneously, ‘response to oxidative stress’ being the highest ranked. The identified mechanism was experimentally validated by using a sensitive and specific bioprobe of mycobacterial cytoplasmic redox potential. The network approach helped to identify most vulnerable targets, which led to the identification of a new drug combination that rescues isoniazid sensitivity in all tested clinical strains, including MDR and XDR strains (Fig. 1). We also tested this approach on trimethoprim-resistant E. coli and here again identified collateral sensitivity through the corresponding network and have successfully tested the strategy with an inhibitor combination (collaboration with Prof. D. Chakraborty, IISc,)
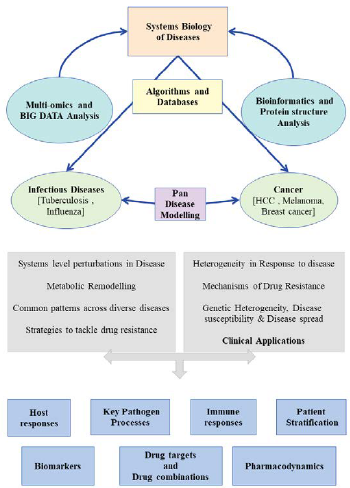
Fig: An overview of the research work in the laboratory. We use a combinations of bioinformatics, systems modelling and experimental approaches to build models of biochemical and cellular processes at multiole levels ranging from individual molecules to whole systems, to address a variety of biomedical questions.
Future Plans
Our next goal is to develop predictive personalized models of susceptibility, disease sub-type and immune status of a patient, so as to strategize more precise therapy. In the next few years, we plan to develop genomics integrated mathematical models to obtain a quantitative appreciation of the precise reaction fluxes involved in the switch with an aim of developing predictive models of estimating risk of active tuberculosis.Given the interest in immune responses and the work we already have about a base human protein-interaction network and HLA genotype profiling, we will investigate cancer specific networks, with the aim of identifying (a) perturbation subnets in selected cancers; using data from patient samples, (we already have preliminary results for melanoma and hepatocellular carcinoma) (b) the ripple effect from the driver mutations to the nodes in the perturbed subnet, and (c) neo-antigens and rank list them based on their affinity to different HLA genotypes and estimate the cytotoxic T-cell responses, and ultimately develop predictive models of disease susceptibility.
- A blood-based host biomarker for discriminating viral and bacterial infections: A clinical decision support tool, Birac-DBT, Grand Challenges India, 2019-2020,
- Development of a general computational framework for systems pharmacology, Department of Biotechnology; 2016-2019;
- Modeling and simulation of cytokine networks; Department of Biotechnology; 2013-2016;
- Exploiting temporal transcription profiles for tuberculosis; Department of Biotechnology;2011-2017;
- Nagarajan, D., Roy, N.,Kulkarni, O., Nanajkar, N., Datey, A., Ravichandran, S., Thakur, C., Sandeep, T., Aprameya, I.V., Sarma, S.P., Chakravortty, D. and Chandra, N. (2019) Ω76: A designed antimicrobial peptide to combat carbapenem-and tigecyclineresistant Acinetobacter baumannii Science Advances, 5(7), p.eaax1946
- Bhagavat, R., Sankar, S., Srinivasan, N. and Chandra,N. (2018) An augmented pocketome: detection and analysis of small-molecule binding pockets in proteins of known 3D structure. Structure. 26,499-512
- Sambaturu, N., Mukherjee, S., López- García, M., Molina-París, C.,Menon, G. I. and Chandra, N. (2018) Role of genetic heterogeneity in determining the epidemiological severity of H1N1 influenza. PLoS Comput. Biol. 14, e1006069
- Nagarajan, D., Nagarajan, T., Roy, N., Kulkarni, O., Ravichandran, S., Mishra, M., Chakravortty, D. and Chandra, N. (2018) Computational antimicrobial peptide design and evaluation against multidrugresistant clinical isolates of bacteria. J. Biol. Chem. 293, 3492-3509
- Sambarey, A., Devaprasad, A., Mohan, A., Ahmed, A., Nayak, S., Swaminathan, S., D’Souza, G., Jesuraj, A., Dhar, C., Babu, S., Vyakarnam, A. and Chandra, N. (2017) Unbiased identification of blood-based biomarkers for pulmonary tuberculosis bymodeling and mining molecular interaction networks. E Bio Medicine. 15, 112-126
Ph.D. Students 9
Undergrads 1
Postdocs 4
Trainees 5