Purusharth I. Rajyaguru

Purusharth I. Rajyaguru
Associate Professor
Ph.D: Centre for Cellular and Molecular Biology, Hyderabad, India
Post-doctoral research:
University of Arizona, Tucson, USA
Year of Joining: 2013
Email: rajyaguru@iisc.ac.in
As a group, we are excited about understanding the movements of mRNA in the cytoplasm between different functional states. These movements contribute to regulation of gene expression at the steps of, (a) Translation and (b) mRNA decay in a variety of cellular processes.
Highlights
- Arginine methylation promotes translation repression activity by promoting interaction with conserved translation initiation factor eIF4G
- Self-association of RGG-motif regulates translation repression activity by competing with eIF4G-binding
mRNA fate decisions play a crucial role in regulating variety of cellular processes including development and differentiation, synaptic plasticity, ageing and diseases such as cancer, neurodegenerative disorders and cystic fibrosis. The proteins associated with mRNA control its individual fate in the cytosol by promoting certain functional transitions and inhibiting others. We aspire to elucidate the molecular mechanisms underlying such movements using Saccharomyces cerevisiae (budding yeast) and human cell culture as model. We are employing a combination of genetic, biochemical, imaging and genomic tools to address the following aims: (a) Understanding the roles of arginine methylation in mRNA fate decisions with a specific focus on RGG-motif proteins; (b) Identifying the mRNA specificity determinants during translation control and decapping; (c) Elucidating the connection between translation control and endocytosis/protein trafficking; (d) Understanding the mechanistic basis of RNA granule disassembly; and (e) Role of translation control and decapping in Cancer establishment and progression.
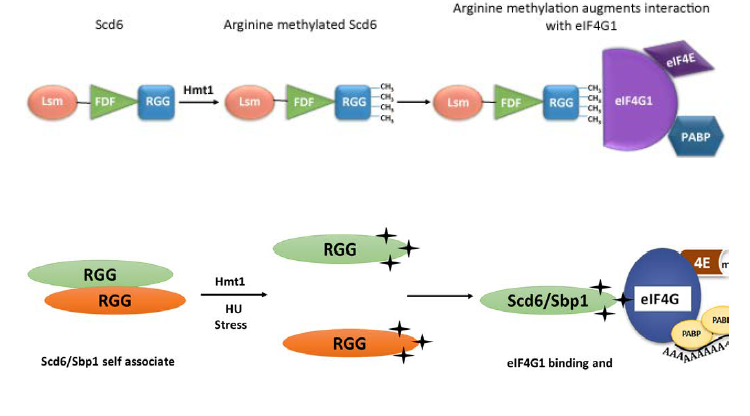
FIG 1 : Arginine methylation of RGG-motif protein translation repressor Scd6 by arginine methyltransferase Hmt1 promotes its repression activity. Methylation augments binding of Scd6 to elF4G1 (Roy and Rajguru,2018,poornima et al., 2019 ).
FIG 2 : Model arguing that self-association of RGG-motif competes with binding to elF4G1. Arginine methylation of Scd6 RGG-motif reduces its ability to self-associate (poornima et al., 2019).
Arginine methylation augments repression activity of Scd6: As a proof-of-concept for aim (a), we have demonstrated that arginine methylation indeed promotes the translation repression activity of RGG-motif containing repressor protein Scd6 by augmenting its interaction with eIF4G. This is the first example of regulation of translation repression by arginine methylation (Fig. 1). Our results were corroborated by another publication that came just a month after our report.
Self-association of RGG-motif competes with eIF4G-binding: It is currently unclear how arginine methylation affects interaction of RGG-domain proteins with eIF4G. Our latest results propose a mechanism whereby RGG-domain self-associates in Scd6 and Sbp1, which compete with RGG-eIF4G interaction. This competition is modulated by arginine methylation. Such modulation appears to be cue-specific since methylation levels go up in response to hydroxyurea treatment. This study proposes a new paradigm in regulation of translation repression through self-association of RGG-domains (Fig. 2).
Future Plans
We aim to understand the role of arginine methylation in regulation of other RGG-motif proteins. Specifically, we would like to understand the mechanism(s) underlying specificty of substrate methylation and role of RNA in determining substrate specificity. RNA granules play an important role in translation control and mRNA decay. One of our research aims to uncover the mechanistic basis of RNA granule disassembly. Through this direction, we hope to understand how mRNAs return back to translation upon removal of stress. We also aim to understand the contribution of RGG-motif proteins to other cellular processes specifically endocytosis and trafficking.
- Understanding mRNA fate decisions: Role of arginine-methylation in functional transitions of RNA-protein complexes (mRNPs); Wellcome Trust-DBT India Alliance Intermediate fellowship; 2013-2018;
- Understanding mRNA fate decisions: Elucidating the role of conserved domains fused to RGG-motifs in translation repression activity of RGG-motif proteins; Department of Biotechnology; 2017-2020;
- Exploring the link between translation control and endocytosis/protein trafficking: Role of translation repressor Scd6 in Clathrin biology; Department of Science & Technology; 2018- 2021;
- Bhatter, N., Roy, R., Shah, S., Sastry, S., Parbin, S., Iyappan, R., Kankaria, S. and Rajyaguru, P. I. (2019) Arginine methylation augments Sbp1 function in translation repression and decapping. FEBS J. 2019. doi: 10.1111/febs.15057
- Poornima, G., Mythili, R., Nag, P., Parbin, S., Verma, P., Hussain, T, and Rajyaguru, P. I. (2019) RGG-motif self-association regulates eIF4G-binding translation repressor protein Scd6. RNA Biol. 2019 Jun 12: 1-13
- Roy, R. and Rajyaguru, P. I. (2018) Stress granules and P-bodies: An insight into mRNA translational control and decay. Proc. Indian Natl. Sci. Acad. 84, 479-491
- Roy, D. and Rajyaguru, P. I. (2018) Suppressor of clathrin deficiency (Scd6) - An emerging RGGmotif translation repressor. Wiley Interdiscip. Rev. RNA: e1479
- Poornima,G., Shah, S., Vignesh, V., Parker, R. and Rajyaguru, P. I. (2016) Arginine methylation promotes translation repression activity of eIF4G-binding protein, Scd6. Nucleic Acids Res. 44, 9358-9368
Ph.D. Students 6
Undergrads 0
Postdocs 1
Trainees 1