Shikha Laloraya

Shikha Laloraya
Professor
Ph.D.: University of Wisconsin-Madison, Madison, USA
Post-doctoral research:
Carnegie Institution of Washington & HHMI, Baltimore, USA
Year of Joining: 2000
Email: slaloraya@iisc.ac.in
My lab investigates chromosome organization and its impact on various chromosomal transactions. Of particular interest are SMC (Structural Maintenance of Chromosomes) complexes that mediate higher order chromatin organization, based on their DNA binding and tethering properties. We also investigate the functional significance of post-translational protein modifications, particularly those mediated by Ubiquitin and SUMO E3-ligases. Recently we investigated roles of a SUMO E3-ligase and a putative ubiquitin-ligase associated with the Smc5/6 complex.
Highlights
- Cohesin modulates gene expression. It is required for SIR-independent repression of subtelomeric genes and for tDNA silencing barrier activity
- Sumoylation mediated by an Smc5/6 complex subunit, Mms21, is crucial for maintenance of chromosome stability
- Interaction of Nse1, a RING-domain protein, with the Smc5/6 complex is required for chromosome replication and stability
The structural organization of chromosomes influences various important chromosomal processes such as transcription, recombination and repair, maintenance of genomic integrity and the accurate inheritance of chromosomes during cell division. Perturbation of chromosome organization can result in defects in these processes, which can manifest as defects in normal growth, development and accumulation of chromosome aberrations, observed in case of disease conditions such as cancer and some developmental disorders. The focus of research of my group is to understand the molecular basis of chromosome organization and explore its relation with these chromosomal processes. We utilize a multifaceted approach including molecular, genetic, cytological and biochemical tools to investigate these processes in the budding yeast Saccharomyces cerevisiae, an excellent model system. Since several aspects of chromosome organization and its regulation are conserved, we plan to extend our findings in yeast to human cell lines, as a step towards exploring the relevance of these findings in understanding human diseases involving chromosomal abnormalities.
Eukaryotic chromosomes undergo extensive structural reorganization in order to facilitate accurate segregation during mitosis. Newly replicated sister chromatids are paired intimately at centromeres and chromosome arms. Chromosomes are also condensed prior to metaphase of mitosis. These structural attributes (cohesion and condensation) are dependent on SMC (Structural Maintenance of Chromosomes) protein complexes, which are evolutionarily conserved, essential for viability and for maintenance of chromosome organization. SMC complexes alter chromosome organization by their ability to bind and bring together different regions of DNA that may be on the same (cis-DNA interaction) or different (trans-DNA interaction) DNA molecule (as shown for cohesin in Fig. 1). Cohesin, Condensin and the Smc5/6 complex are three well-known SMC complexes of interest to us.
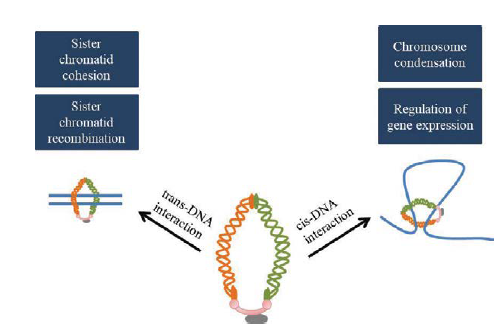
FIG 1: Cis and trans-DNA interactions of the cohesin complex. Trans DNA interaction is important for cohesin's role in sister chromatid cohesion and sister chromatid recombination, Wheresas cis DNA interactions resulting in formation of cohesin mediated loops, are important for chromosome condensation and regulation of gene expression.
Cohesin is a conserved SMC complex, first identified due to its role in sister chromatid cohesion. Dysfunction of cohesion proteins results in disorders in humans characterized by severe developmental abnormalities, and is also associated with cancer, highlighting the biomedical relevance of functional studies of Smc proteins. As a step towards understanding the molecular basis of cohesion, I had earlier identified the chromosomal addresses of Mcd1p, a component of cohesin, leading to identification of specific cohesin binding sites or Cohesin Associated Regions (CARs) on yeast chromosomes. Our group also identified a cohesin dependent silencing boundary element that can limit the spread of rDNA (ribosomal DNA) silencing, suggesting that cohesin can affect gene expression.
Since cohesin dysfunction associated alterations in gene expression may result in developmental defects observed in disorders caused by cohesin deficiency, our current focus is to understand additional roles of cohesin in gene expression. We recently uncovered a role for cohesin in repression of subtelomeric gene expression that is independent of the SIR complex, a well-known regulator of silencing in budding yeast. Silencing of gene expression in extensive subtelomeric regions is SIR-independent and poorly understood. We found that clusters of subtelomeric genes are preferentially derepressed in cohesin mutants while SIR binding was unaltered. Our findings reveal that cohesin dependent chromatin organization may be important for silencing of genes in extended subtelomeric domains.
We also study the Smc5/6 complex that is required for recovery from replication stress and DNA repair. Recently, we have investigated the role of Mms21/ Nse2, one of the subunits of the Smc5/6 complex that has SUMO E3-ligase activity and hence can conjugate SUMO (Small Ubiqitin related Modifier) to various protein targets. By creation of SUMO- ligase defective mutants of Mms21 and sumoylation defective variants of its targets, we are testing the importance of sumoylation of chromosomal protein targets of Mms21. We found that Mms21 mediated sumoylation is crucial for the maintenance of chromosome stability, especially of torsionally constrained chromosomes. We also study Nse1, a RING-domain subunit of this complex is a putative ubiquitin ligase required for chromosome replication and stability. We are currently investigating the role of the Nse1, 3 and 4 trimeric subcomplex of the Smc5/6 complex.
- Science and Engineering Research Board (SERB), INDIA
- Council of Scientific and Industrial Research (CSIR), INDIA
- Department of Biotechnology (DBT), INDIA
- The Wellcome Trust (WT), U.K.
- Kothiwal, D. and Laloraya, S. (2019) A SIR independent role for cohesin in subtelomeric silencing and organization. Proc Natl Acad Sci U.S.A. 116 (12): 5659-5664
- Laloraya, S. (2018) Asymmetric tyrosination of spindle microtubules facilitates selfish inheritance. Trends Cell Biol. 28, 417-419
- Wani, S., Maharshi, N., Kothiwal, D., Mahendrawada, L., Kalaivani, R., Laloraya, S. (2018) Interaction of the Saccharomyces cerevisiae RING-domain protein Nse1 with Nse3 and the Smc5/6 complex is required for chromosome replication and stability. Curr. Genet. 64, 599-617
- Rai, R., Varma, S. P., Shinde, N., Ghosh, S., Kumaran, S. P., Skariah, G. and Laloraya, S. (2011) Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J. Biol. Chem. 286, 14516-14530
- Biswas, M., Maqani, N., Rai, R., Kumaran, S. P., Iyer, K. R., Sendinc, E., Smith, J. S. and Laloraya, S. (2009) Limiting the extent of the RDN1 heterochromatin domain by a silencing barrier and Sir2 protein levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 29, 2889-2898
Ph.D. Students 5
Undergrads 4
Postdocs 2
Trainees 0